[News: Someone linked this text from the Open Source Toolkit: Hardware article collection and channel of PLoS. Thanks.]
I built my first fluorescent microscope. A Leitz Labovert became a surprisingly decent fluorescent microscope for DIY after spending about £100 and 3D printing a few custom parts. It is a simplified design with only a barrier and an excitation filter, LED illumination and no dichroic mirror.
The quality is sufficient for observing fluorescent yeast and in general fairly faint Drosophila embryos in low-to-midrange magnification.
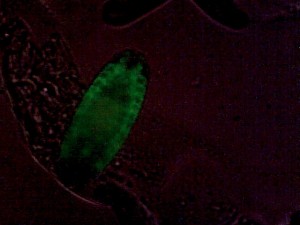
Drosophila embryonic tracheal system, GFP-marked. Weak additional bright-field illumination for embryo outline.
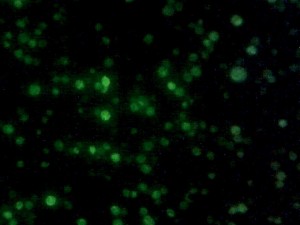
Yeast with nuclear GFP marker.
Fluorescence is short wavelength light in, longer wavelength light out. In principle a fluorescent microscope needs only two parts: 1) a light source that emits light at the excitation, but not at the emission wavelengths of the sample, and 2) a barrier filter, which lets only the emitted longer wavelength light through, so it doesn’t drown out in the excitation light. There are a few details though, and I will start with the barrier filter.
Continue reading →
![David Molnar [Update:, PhD]](http://www.srcf.ucam.org/~dm516/wp-content/themes/twentyeleven/images/headers/chessboard.jpg)

